Research
Evolutionary dynamics of genomes and epigenomes
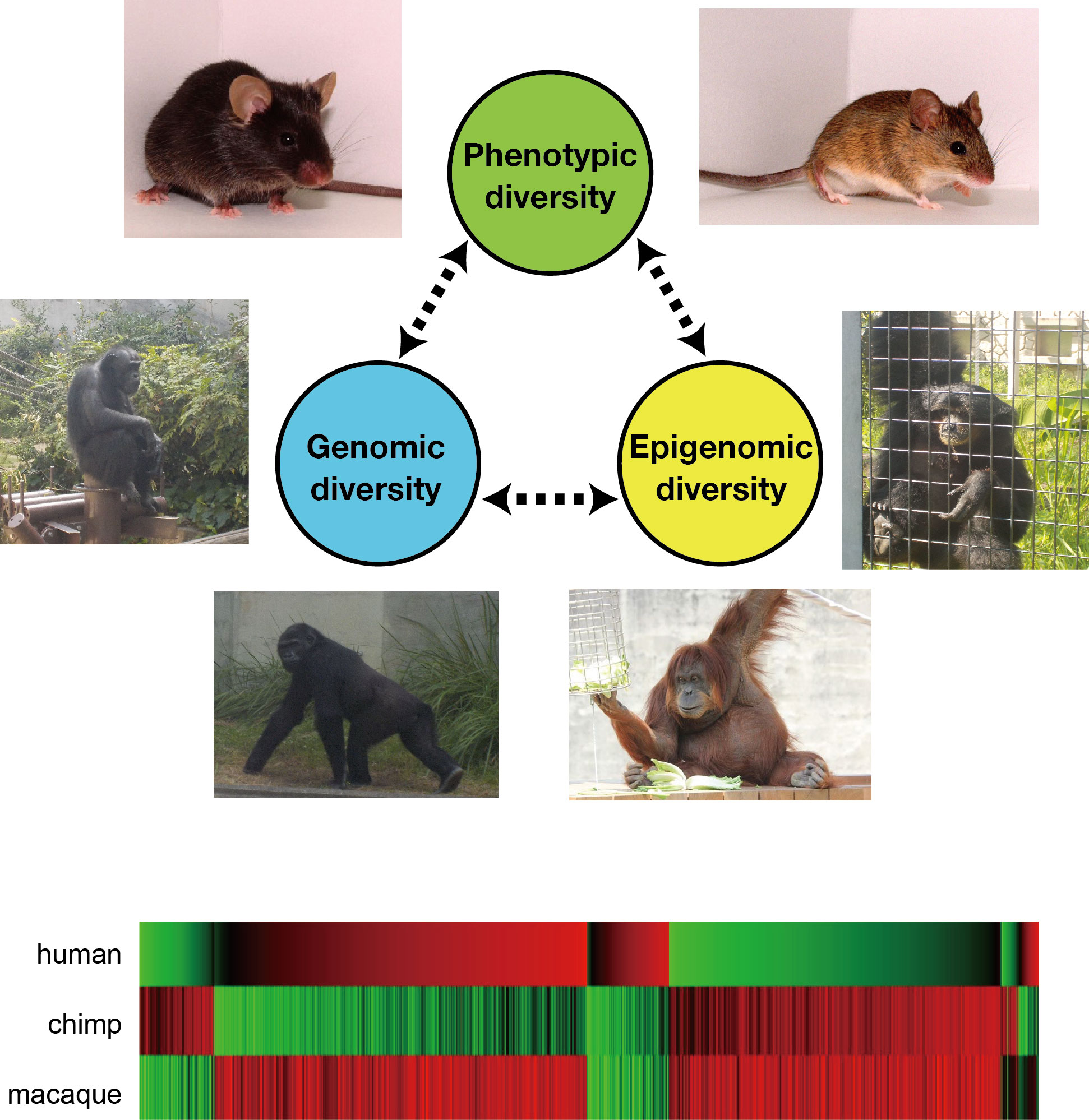
Cells in an individual have the exactly same genomic sequence in principle. But the genome-encoded functions are decoded in different regions in different cell types, which underlies cell-type-specific gene expression profiles. This regulatory mechanism is called epigenetics, and achieved mainly by chemical modifications (e.g., methylation) of genomic DNA and histone proteins. Epigenome is the whole of epigenetic modifications present in a cell.
Epigenetic diversity among individuals sometimes generates phenotypic diversity. We aim to understand the molecular mechanisms that generate epigenetic differences and how genome and epigenome affect each other during evolution. We study the genomic and epigenomic diversities within a species (mouse) and between species (primates) by using next generation sequencer.
Functions of retrotransposons and their regulation
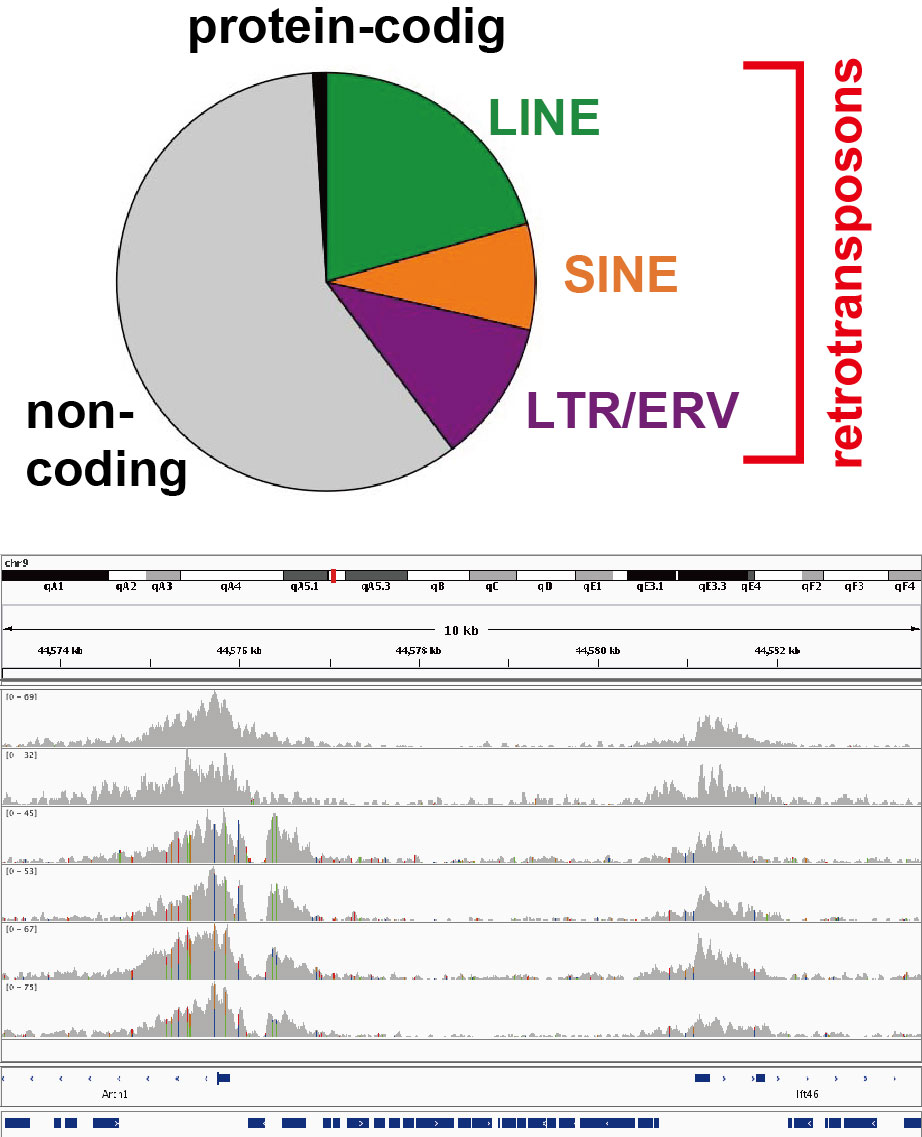
In mammals including humans, protein-coding sequences make up only 1% of the genome. About a half of the rest are occupied by retrotransposons, a class of mobile genetic elements. Although retrotransposons were regarded as junk DNA, they are now recognized as a driver of genome evolution. Moreover, evidence has accumulated that retrotransposons can regulate other parts of the genome in various ways. We study such retrotransposon-encoded functions, their regulation, and the effects of retrotransposon transposition on the host epigenome.
Genomic integrity of germ cells
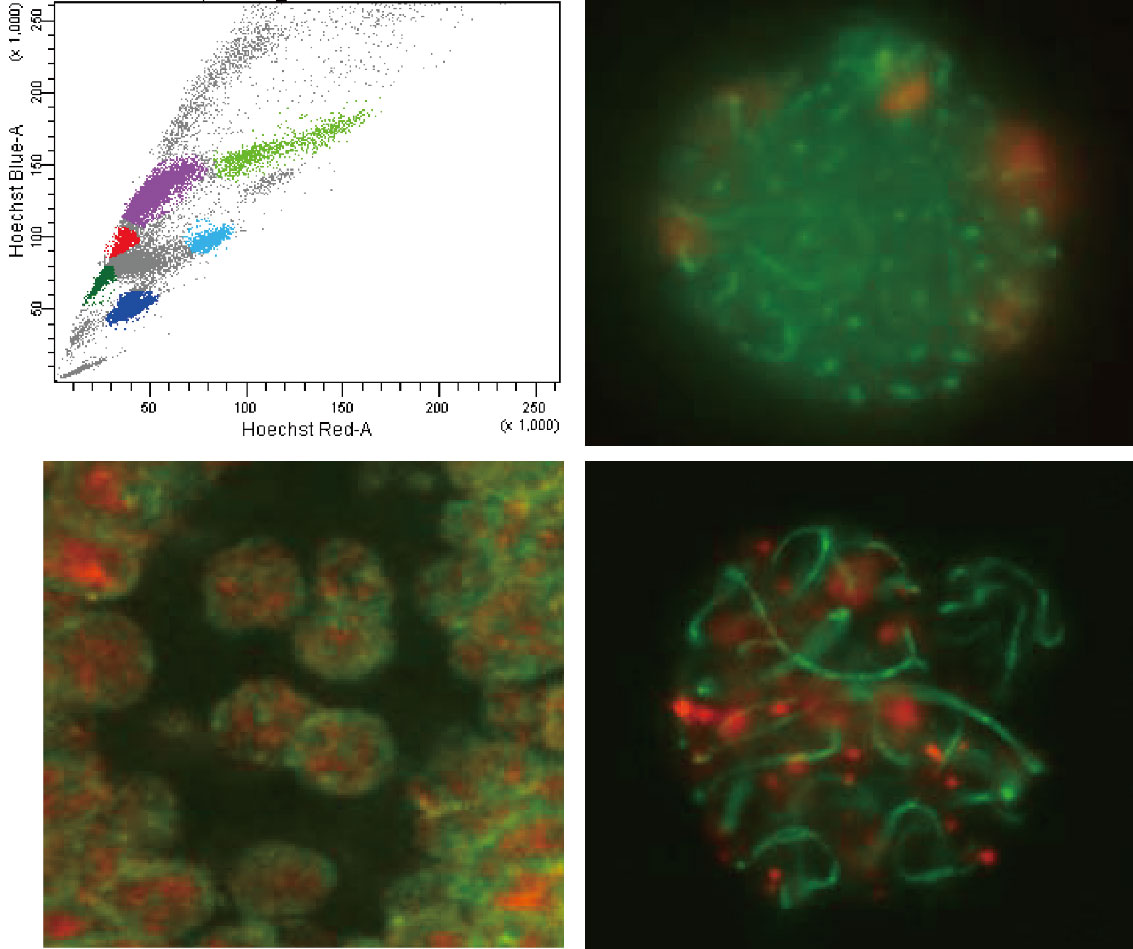
Germ cells are the sole type of cells that convey the genome information to the next generation. Thus, they must maintain the genomic integrity by suppressing mutagenic events, such as retrotransposon transposition and unequal crossing over in meiotic homologous DNA recombination. We study how the retrotransposons are epigenetically silenced during germ cell development and how meiotic recombination events are epigenetically regulated.
Transgenerational epigenetic inheritance
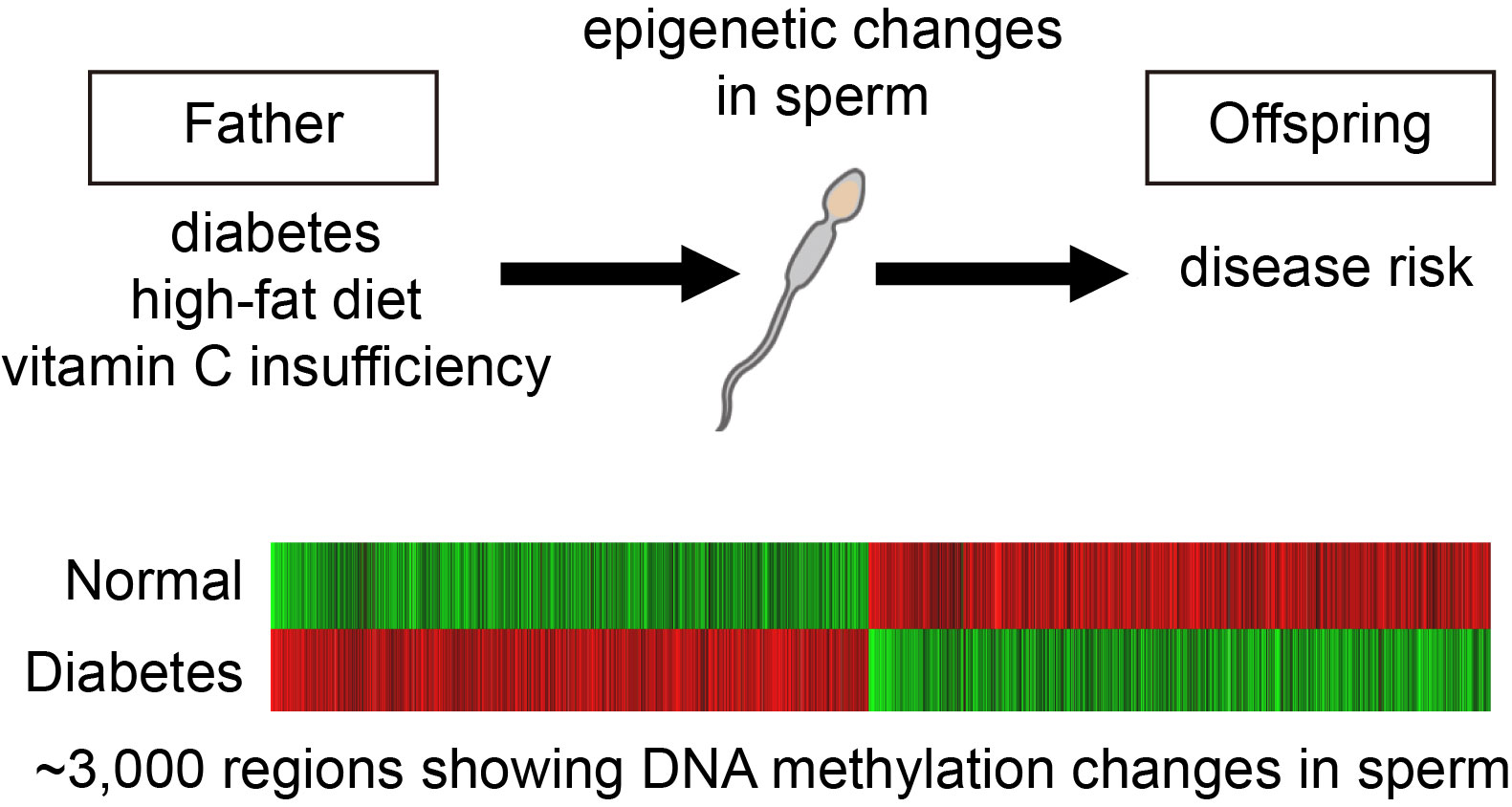
Why traits are inherited from parent to child is because the child receives the DNA of the same base sequence as the parent; that is the genetic mechanism of inheritance. In recent years, however, it has started to be thought that, in addition to DNA base sequences, other types of genetic information may be transmitted in mammals . Indeed, in plants, DNA methylation status is known to be inherited by offspring. In mammals, it is possible that the health and nutritional status of the parents, such as diabetes or a high-fat diet, could affect the offspring born later in their life, even though their genome is not mutated. Our lab has worked to elucidate the molecular mechanisms and how it is transmitted to the offspring. For example, we have found that diabetes changes the methylation state of sperm DNA. This is one of the most promising candidates for epigenetic information to be transmitted to the offspring.
Epigenetic sex determination system in mealybug

In humans, a pair of sex chromosomes determines the sex, in which females have two X chromosomes whereas males have both X and Y chromosomes. Which combination of the sex chromosomes an individual has is genetically determined at the time of fertilization. Some animals, however, do not have sex chromosomes. The mealybug has five pairs of autosomes and no sex chromosome, thereby females and males have the same set of chromosomes. In females, all 10 chromosomes are in an active state. On the other hand, when a set of chromosomes are epigenetically inactivated early in development, that individual becomes a male. Once inactivated, the chromosomes remain inactive during the lifetime. All inactivated chromosomes are inherited from the father. We have studied molecular mechanisms involved in the epigenetic sex determination.



